 |
BHAC Python tools
|
 |
BHAC Python tools
|
This document describes the use of Python for plotting BHAC 1D and 2D simulation data. The Python tools were initiated by Oliver Porth.
The Python macros are located in the directory tools/python and depend on VTK-bindings and the widespread SciPy toolkit.
If you are a beginner at Python, we recommend using a pre-configured python-distribution such as Enthought Canopy. Note that for VTK support you will need to sign up for the free academic version. This distribution comes with SciPy which bundles essential numerical tools (numpy), a plotting backend (matplotlib) and a command-line interface called IPython for interactive use.
Often, a simple
does the trick.
You can verify your VTK installation by trying to import vtk in a Python session. If VTK is installed properly, this should throw no error messages.
The tools are installed by adding the directory $BHAC_DIR/tools/python to your python path. This can be done in a number of ways. Either you expand your $PYTHONPATH environment variable, e.g. on the bash shell:
or you add a file userpath.pth in one of the folders existing in the search path already. The file simply contains
where youramrvacdir has to be the resolved variable $BHAC_DIR, e.g. /Users/Max/code/amrvac. For example on a mac, you can add the userpath.pth to the directory /Library/Frameworks/Python.framework/Versions/Current/lib/python2.7/site-packages or similar.
We currently support reading the cell-centered VTK file-types .vtu, .pvtu, .vti as well as binary particle .dat files and some ascii .csv files for Python. This means that you can use the same files with e.g. Paraview or Visit. These can be created in parallel and during runtime (see also the section on data-file conversion). Reading data is handled in the file ready.py which provides various classes for different data.
For interactive use with the terminal, cd to the directory containing your data and fire up ipython:
Import the reader and plotter classes:
This uses the class read.load.
Interactively load the data:
which loads data'offset'.vtu, where offset is zero-padded to four digits (e.g. 0010). The data is contained in the instance d. Thus d.rho holds the numpy-array of the density, d.v1 hols the first component of velocity (assuming your data is in primitive quantities and your velocity variable is called 'v1') and so on. To load a different snapshot into the instance e, type
In order to load a file ending in .pvtu, change the attribute of the optional parameter type to type='pvtu'. The default is currently type='vtu'.
This uses the class read.loadvti. For uniform .vti files, you can use
This uses the class read.particles. Type
to import the particle snapshot data into python.
This uses the class read.ensemble to load particle ensemble comma separated value files. Type
to load file data0000_ensembleoffset.csv where offset is zero padded to six digits. npayload stands for the number of payload arrays in your data (default is one). We have two running indices in the output files, where the first data0000 describes the fluid snapshot (e.g. useful when adding particles to a frozen snaphot) and the second offset denotes the particle ensemble snapshot number.
To load a 1D slice in .csv format from a 2D simulation, you can use the class read.loadcsv
which loads file data_ddir_xcoord_noffset.csv, e.g. data_d1_x+0.00D00_n0000.csv.
Once the data is loaded into the instance d, you can access it with d.varname where varname is a variable name chosen in your AMRVAC parameter-file. Pythons powerful introspectrion makes it real easy to see whats available in the data: just type d. and hit the tab key to see whats available. For uniform data (.vti), the arithmetic center of the cells can be obtained via the command
see read.load.getCenterPoints for implemetation details. Thus you can make a simple 1D plot using matplotlib
in solid ('-') black ('k') with '+' symbols. At this point d.getCenterPoints() and d.rho are simply numpy arrays so you can use all of matplotlib for your plotting experience.
For the structured .vti files, the attribute read.loadvti.x (or .y or .z) denotes the cell center coordinate, such that a plot would be obtained with
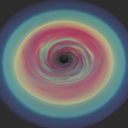
The module amrplot contains plotting tools for 2D AMR data. These come in two flavours: the class amrplot.polyplot to show each cell in one patch and the class amrplot.rgplot to show data interpolated onto a uniform mesh. Other than that, both classes offer the same functionality. To display a 2D image of the density in your datastructure d, invoke a new instance
which opens a new window containing your data. Drawing a window for the first time might take a few seconds as a list of the cell patches has to be generated. You can re-plot another quantity (e.g. temperature ~p/rho) in this window with the command
amrplot.polyplot has several convenient members, for example you can save your image to a file using amrplot.polyplot.save which takes the filename as a parameter. The AMR blocks can be displayed by setting the attribute
which can be switched off by setting amrplot.polyplot.blocks back to None. You can fix the axis-range with the attributes amrplot.polyplot.xrange and amrplot.polyplot.yrange and fix the window zoom by setting amrplot.polyplot.fixzoom to a value other than None.
The method amrplot.polyplot.onkey allows you to inspect a location in your data: use the middle mouse button to mark a cell in your plotting window, then a pink crosshair will be displayed and the flow variables at chosen location are returned to the terminal.
Some auxiliary functions you might find useful:
This documentation is created using doxypypy and the documentation system Doxygen. Simply run
in the $BHAC_DIR/tools/python folder to re-generate the documentation.
 1.8.13
1.8.13